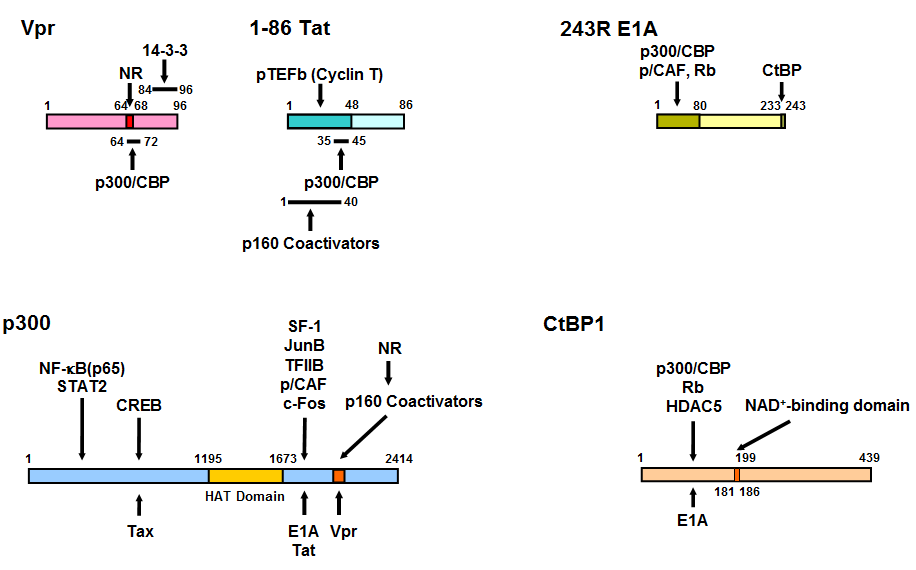
Figure 17: Linearized Vpr, Tat, E1A, p300 and CtBP1 molecules and their mutual interaction domains.
Vpr interacts with GR through its LXXLL motif located from amino acid 64 to 69. Binding sites of p160 nuclear receptor coactivators and Vpr to p300 overlap with each other and they both bind NRs. Thus, Vpr mimics the host p160 nuclear receptor coactivators and enhances NR transcriptional activity. Tat also binds both p300 and p160 nuclear receptor coactivators. p300 facilitates attraction of transcription factors, cofactors and general transcription complexes by loosening the histone/DNA interaction through acetylation of histone tails by its histone acetyltransferase (HAT) domain. E1A binds p300 at the latter’s C-terminal portion, while it physically associates with the N-terminal portion of CtBP1 through its C-terminal end. The N-terminal portion of CtBP1 physically interacts with HDAC5 and Rb, which have repressive activity on transcription. CtBP1 regulates interaction with its binding partners by sensing NADH levels through its NAD+-binding domain. The HAT domain of p300 and the NAD+-binding domain of CtBP1 are indicated in grey. Modified from (365).
CREB: CRE-binding protein, HAT: histone acetyltransferase, HDAC5: histone deacetylases 5, NF-kB: nuclear factor-kB, NAD: nicotinamide adenine dinucleotide, NR: nuclear hormone receptor, p/CAF: p300/CBP-associating factor, pTEFb: positive-acting transcription elongation factor b, Rb: retinoblastoma protein, SF-1: steroidogenic factor-1, STAT2: signal transducer and activator of transcription 2, TFIIB: transcription factor IIB.
